Whole-Transcriptome RNA-Seq Data Analysis using Linux
Why take this course?
🌱 Dive into the World of Genomics: Whole-Transcriptome RNA-Seq Data Analysis using Linux
🚀 Course Headline: Unlock the Secrets of Gene Expression with Whole-Transcriptome RNA-Seq Data Analysis using Linux – A Journey into Bioinformatics with Dr. Rashid Saif!
🔍 Course Description: Are you ready to elevate your bioinformatics skills to the next level? This comprehensive course, "Differential Gene Expression Analysis," is tailored for students and professionals navigating the intersection of molecular genetics and bioinformatics. Dr. Rashid Saif will guide you through the intricacies of measuring differential gene expression using whole-transcriptome RNA-Seq datasets.
🔍 What You Will Learn:
- The significance of computational skills in life sciences research.
- The types of biological data amenable to computation and statistical analysis.
- How Linux/R can be harnessed to address multi-omics problems.
- The evolution of bioinformatics and computational biology to meet modern genetics challenges.
- The transition from a wet lab environment to a dry lab setting through bioinformatics data science.
- Mastery of bioinformatics tools for analysis, visualization, and interpreting your large transcriptomic datasets.
👨🔬 Course Highlights:
- Hands-On Experience: Engage with real-world RNA-Seq datasets to perform differential gene expression analysis.
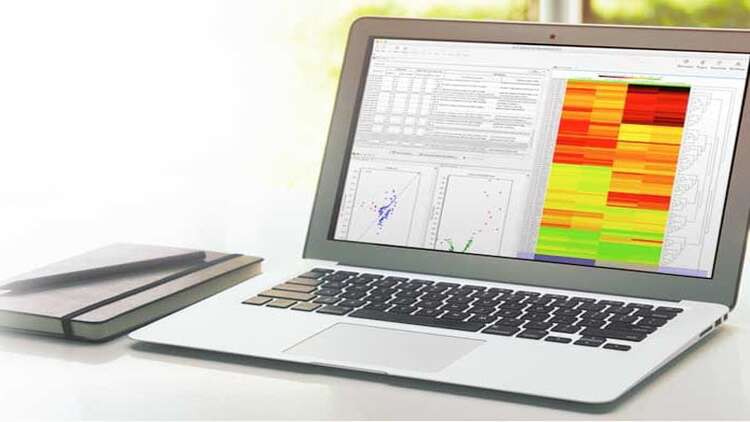
- Skill Development: Learn bash scripting in Linux CLI and apply R statistical packages for robust data visualization, resulting in comprehensive heatmap graphs.
- Resource Optimization: Analyze multi-omics data efficiently even with limited financial resources and without extensive programming knowledge.
🌟 Who Should Take This Course? This course is perfect for:
- Students pursuing degrees in genomics, bioinformatics, or related fields.
- Researchers looking to expand their skillset into computational biology.
- Professionals working in biotechnology companies or pharmaceutical firms.
- Anyone interested in understanding the impact of gene expression on biological processes.
🔬 Course Outline:
- Introduction to RNA-Seq and its role in genomics research.
- Setting up your Linux environment for bioinformatics analysis.
- Quality control and alignment of raw sequencing reads to reference genomes.
- Quantification of gene expression levels using tools like featureCounts or HTSeq.
- Statistical modeling and differential expression analysis with DESeq2, limma, or edgeR.
- Data visualization through heatmaps and principal component analysis (PCA).
- Best practices for data management and reproducibility in bioinformatics.
📊 Key Takeaways:
- A deeper understanding of differential gene expression analysis.
- Proficiency in using Linux and R for handling and analyzing NGS data.
- Ability to visualize and interpret complex biological datasets effectively.
- Insight into the future landscape of life sciences research with a focus on computational methods.
By the end of this course, you'll not only be equipped with the skills to conduct differential gene expression analysis but also be prepared to transition into a more data-centric role within the realm of genetics and biology. Join us, transform your skillset, and become a bioinformatics expert! 🌱✨
Course Gallery
Loading charts...