Learn Molecular Dynamics from Scratch
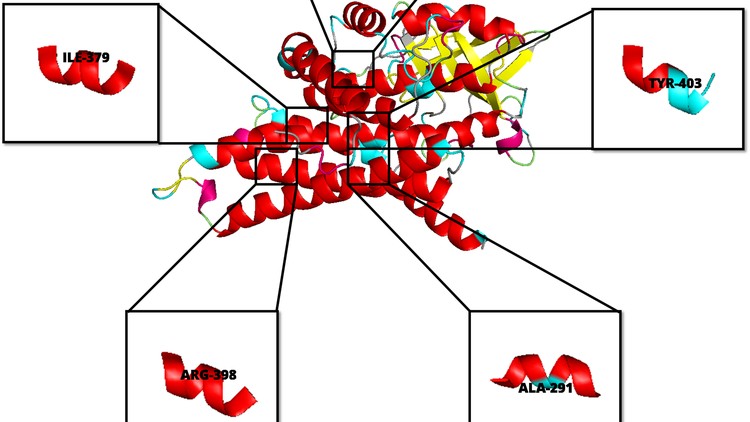
Why take this course?
Unlock the Secrets of Molecular Dynamics with GROMACS: A Hands-On Journey 🧬✨
Are you ready to dive into the dynamic world of molecules? Whether you're a student, researcher, or simply a science enthusiast, "Learn Molecular Dynamics from Scratch" course is your gateway to understanding the experimental approach using GROMACS. Led by our expert instructor, Thirumal Kumar, this comprehensive online course will take you through the intricacies of molecular dynamics simulations without overwhelming you with complex theories.
Course Highlights:
- 🎓 Comprehensive Introduction: Understand what Molecular Dynamics (MD) is and why it's a powerful tool in modern science.
- 🧪 Force Fields: Learn about the force fields that govern molecular interactions and how to choose the right one for your simulations.
- Software Mastery: Get hands-on experience with MD software, focusing on GROMACS – one of the most popular and versatile tools available.
- 🤿 Ubuntu & Windows: Navigate through the Ubuntu environment, even if you're a Windows user at heart, to set up your workstation for MD simulations.
- 📚 GROMACS Installation and Usage: Master the installation process and learn to manipulate GROMACS files with ease.
- 🧫 Molecule Preparation: Discover the steps to prepare your molecular system for simulation, ensuring accuracy and efficiency.
- 🔨 Energy Minimization: Learn techniques to remove high-energy structures from your model before proceeding to equilibration.
- ⚖️ System Equilibration: Achieve an equilibrium state for your system, preparing it for the dynamic simulations ahead.
- 🚀 Molecular Dynamics Simulations: Run MD simulations and interpret the results in a meaningful way.
- 🔍 Trajectory Analysis: Analyze the trajectories from your simulations to extract valuable insights into molecular behavior and dynamics.
What You Will Learn:
- The fundamental principles of molecular dynamics
- How to set up and run MD simulations with GROMACS
- Best practices for molecular preparation, energy minimization, and equilibration
- Techniques for analyzing complex biological systems and molecular interactions
- Tips and tricks for optimizing your simulations and interpreting the results
Why Choose This Course?
- Practical Approach: We believe in learning by doing. You'll perform real simulations and analyze real data.
- Expert Guidance: Our instructor, Thirumal Kumar, has extensive experience in molecular dynamics and will provide valuable insights throughout your learning journey.
- Flexible Learning: Access course materials at your own pace, from the comfort of your home or office.
- Community Support: Join a community of like-minded individuals and engage with peers through forums and discussions.
Ready to Embark on This Exciting Adventure? 🚀🧚♂️
Enroll in "Learn Molecular Dynamics from Scratch" today and transform your understanding of molecular interactions and dynamics. With GROMACS as your toolkit, the world of computational biophysics is yours to explore! 🎉
Join us now and take the first step towards mastering the science of motion at the molecular level. Let's embark on this journey together and unveil the mysteries of life itself! 🌟
Course Gallery

Loading charts...